Robust Registration of Statistical Shape Models for Unsupervised Pathology Annotation
Published at: LABELS: International Workshop on Large-scale Annotation of Biomedical data and Expert Label Synthesis, 2019[ PDF ] [ POSTER ]
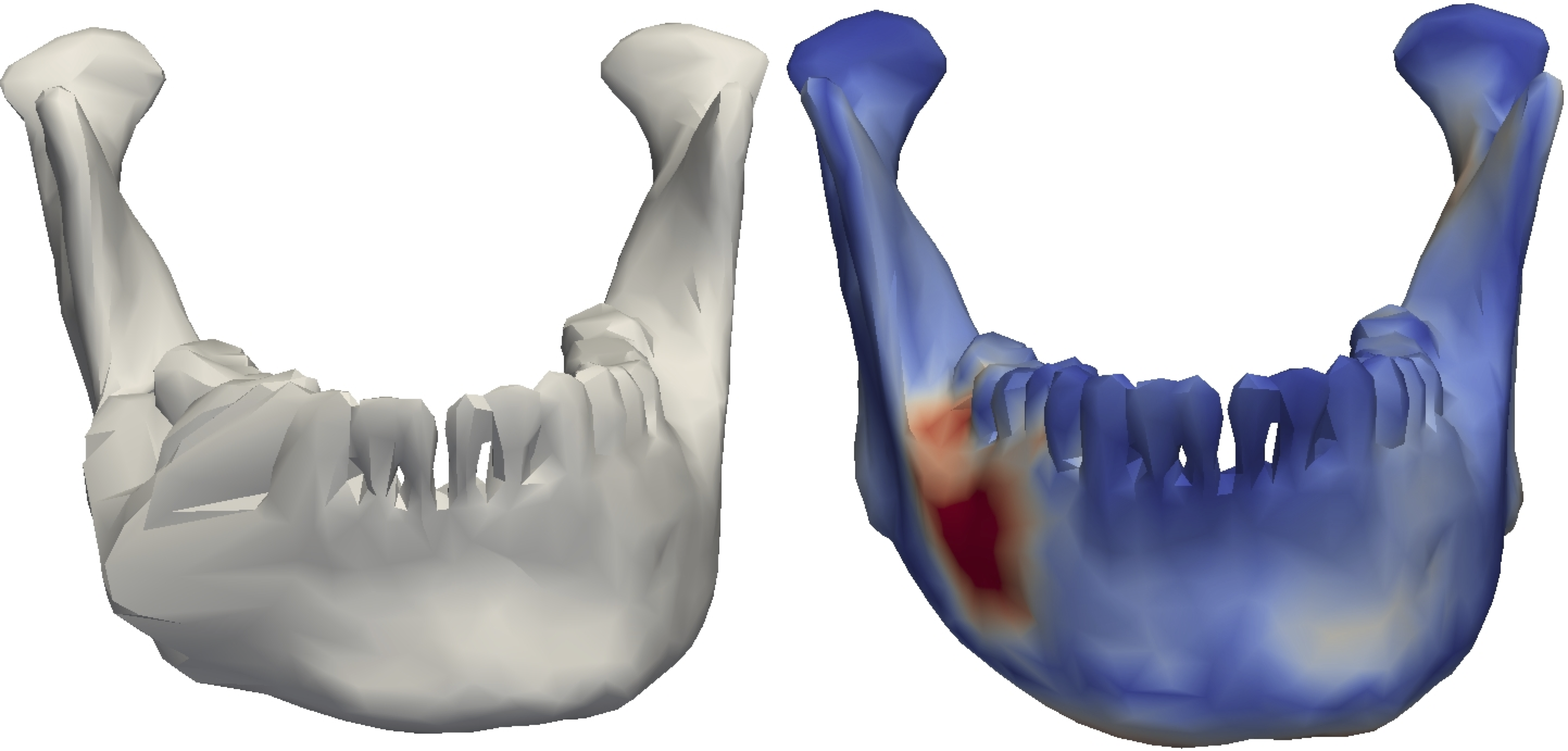
Abstract
We present a method to automatically label pathologies in volumetric medical data. Our solution makes use of a healthy statistical shape model to label pathologies in novel targets during model fitting. We achieve this using an EM algorithm: the E-step classifies surface points into pathological or healthy classes based on outliers in predicted correspondences, while the M-step performs probabilistic fitting of the statistical shape model to the healthy region. Our method is indepen- dent of pathology type or target anatomy, and can therefore be used for labeling different types of data. The method is able to detect pathologies with higher accuracy than standard robust detection algorithms, which we show using true positive rate and F1 scores. Furthermore, the method provides an estimate of the uncertainty of the synthesized label. The de- tection also directly improves surface reconstruction results, as shown by a decrease in the average and Hausdorff distances to ground truth. The method can be used for automated diagnosis or as a pre-processing step to accurately label large amounts of images.
